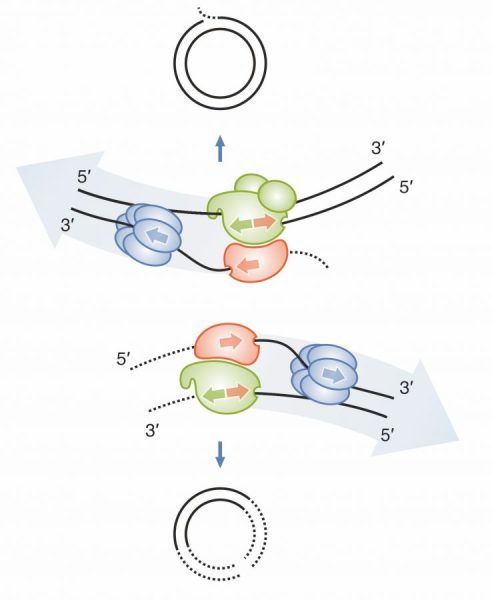
If you want to know what happens when the circular mitochondrial genome breaks and suddenly has two ends (aka linear), take a look at our new paper in Nature Communications. Would you ever believe that exonucleases famous for their fine sculpting abilities could be so brutal?
Archive for the ‘mtDNA’ Category
When the wheel breaks
May 1, 2018Link to MitoWheel
May 12, 2016If you have a website dealing with human mitochondrial DNA and you would like to show specific positions graphically, you can now make a direct link to MitoWheel. Simply send your query through the HTML address line appending a question mark and the query text. Something like this:
http://mitowheel.org/mitowheel.html?4277C
or
http://mitowheel.org/mitowheel.html?tRNALeu
Small addition: using the left and right arrow keys on your keyboard, you can now walk through the sequence nucleotide by nucleotide. If you want to make bigger jumps, type into the search field ‘+42’ or ‘-17’ and press ENTER repeatedly.
MitoWheel 2.0
April 5, 2016MitoWheel was updated on April 5th, 2016.
![]() Well, it is actually more than just an update. MitoWheel has been re-made from scratch and is now fully based on Java. This means that you will be able to use it on mobile devices or some smart TVs. The actual version contains all features that I myself regularly use. Nevertheless, some functions of the old version are not yet implemented. If you cannot do without them, you will find a link on the ‘About’ page to the predecessor. This you can access by clicking on the ‘+’ sign. New is the orange button in the search field that serves for launching a query or walking through multiple matches, things that otherwise can be done by pressing ENTER. On the other end of the search field, you will find the search icon, similar to the old version. New is that you can click on it to load files with lists of queries, most importantly with lists of SNPs. The files should be simple text files in FASTA format (‘>Sequence_Name’ followed by a comma-separated list of SNPs in one or more lines) or VCF-type text files that are used to represent exome sequencing data. If you would like to use file types that do not load into MitoWheel 2.0, email me a sample, and I will do my best to extend its file reading capabilities.
Well, it is actually more than just an update. MitoWheel has been re-made from scratch and is now fully based on Java. This means that you will be able to use it on mobile devices or some smart TVs. The actual version contains all features that I myself regularly use. Nevertheless, some functions of the old version are not yet implemented. If you cannot do without them, you will find a link on the ‘About’ page to the predecessor. This you can access by clicking on the ‘+’ sign. New is the orange button in the search field that serves for launching a query or walking through multiple matches, things that otherwise can be done by pressing ENTER. On the other end of the search field, you will find the search icon, similar to the old version. New is that you can click on it to load files with lists of queries, most importantly with lists of SNPs. The files should be simple text files in FASTA format (‘>Sequence_Name’ followed by a comma-separated list of SNPs in one or more lines) or VCF-type text files that are used to represent exome sequencing data. If you would like to use file types that do not load into MitoWheel 2.0, email me a sample, and I will do my best to extend its file reading capabilities.
Short summary of features:
- Navigate by clicking on the point of interest either on the sequence bar or on the Wheel
- If you are bored, you can just flick the sequence bar or the Wheel. You have won if it stopped in the anticodon of a tRNA (chances are 66 to 16569)
- Search for nucleotide sequence motifs, now including ambivalent positions. You can either use usual IUPAC nucleotide codes (‘R’ for ‘A’ or ‘G’, ‘Y’ for ‘C’ or ‘T’, etc.) or indicate the alternative nucleotides in square brackets (‘AC[GC][AGCT]CC[AT]GC’)
- Search for terms like ‘protein’, ‘MT-TS2’, ‘complex IV’
- If you want to search for multiple terms, separate them by comma
- Load files by clicking on the search icon
- The purple frequency bars, located below or above each nucleotide, are now calculated based on 30,181 human database sequences. Exact numbers are displayed if you search for specific alleles (‘5846C’ or ‘m.12367A>G’)
Note on privacy: Once loaded into your browser, MitoWheel 2.0 functions entirely offline. No data are sent through the net. In case some future functions will require a connection to the server, this will be explicitly indicated.
I hope you will have fun with MitoWheel 2.0. If you miss some features, either from the old version or something that just crossed your mind, do not hesitate to email me. MitoWheel is probably not free of errors. If you notice one, please let me know. Just as in the last eight years, keep the Wheel rolling!
MitoWheel: The Manual
April 6, 2008The MitoWheel was updated on April 6th, 2008.
There were small corrections made to the program. However, the most relevant change happened to the web site. I deleted the words ‘Under construction’ in the section ‘How to use’, and I wrote quite a lot of other words instead. Most of the instructions and tricks have been already mentioned here in the blog, but now you can find a more systematic guide right there where the Wheel is. To make the manual more enjoyable, I also included three short videos showing the Wheel in action.
As an appetizer, here you can see the first video about navigating the Wheel. For better quality, check out the original.
A Little Make-Up
March 30, 2008The MitoWheel was updated on March 30th, 2008. This time new:
- After a few years of hesitation, I finally switched the orientations of the two replication origins. If you prefer the previous version, or you find some evidence that the previous version was more correct, please let me know.
- Now you can really spin the Wheel directly! Just click on it and drag it around.
Sequence Bar with Allele Frequencies
March 23, 2008The MitoWheel was updated on 23th March, 2008. This time new:
The sequence bar now contains information not only about the function of specific nucleotides, but also about allele frequencies at polymorphic positions. Each little gray bar above or below a nucleotide letter represents the number of individuals who carry a difference (also known as SNP) at the given position as compared to the revised Cambridge reference sequence. To be able to show both very rare and very frequent polymorphisms, the bars have a logarithmic scale. I hope you will find this new feature useful. This can help you to design reliable PCR primers for the human mitochondrial genome. After all, you don’t want your primer’s 3′-end sitting right on a very frequent polymorphism (risking that under certain conditions you will not be able to amplify a PCR fragment from a subset of individuals). The allele frequency bars are also good starting points if you want to explore the diversity of the human population by using the “+” and “-” operators in the search box (as described here).
I hope you will find this new feature useful. This can help you to design reliable PCR primers for the human mitochondrial genome. After all, you don’t want your primer’s 3′-end sitting right on a very frequent polymorphism (risking that under certain conditions you will not be able to amplify a PCR fragment from a subset of individuals). The allele frequency bars are also good starting points if you want to explore the diversity of the human population by using the “+” and “-” operators in the search box (as described here).